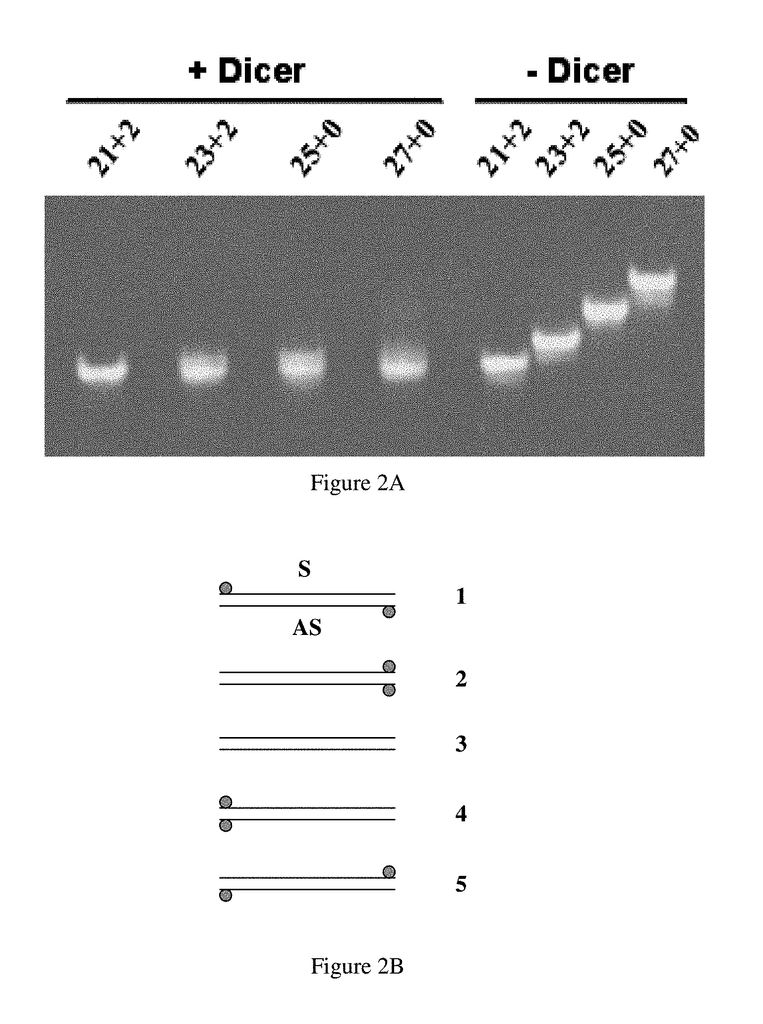
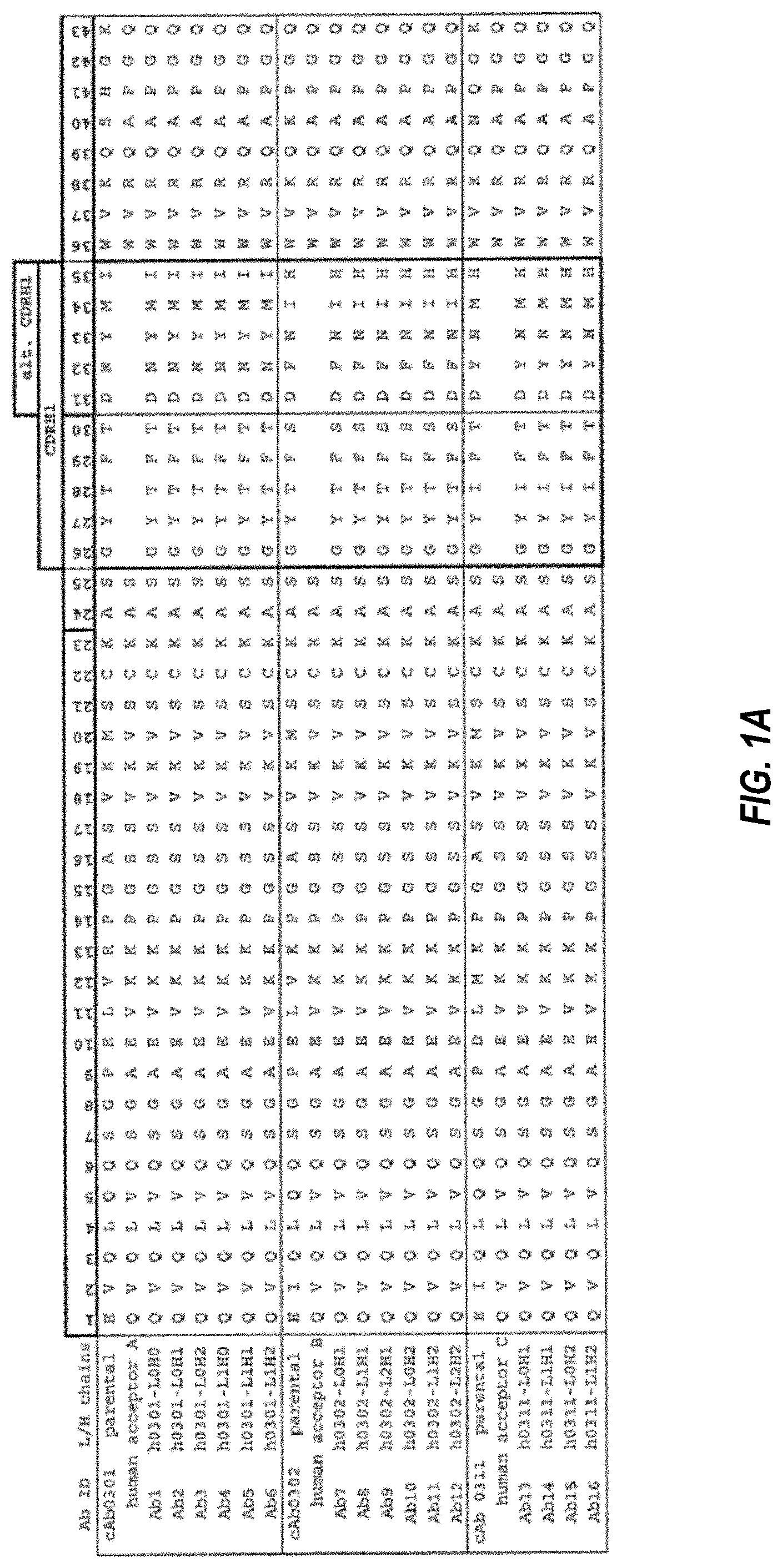
Invented by John J Rossi, Mark A. Behlke, Dongho Kim, Integrated DNA Technologies Inc, City of Hope
Inhibition of gene expression has been a focus of research for many years, and there are several methods and compounds that have been developed to achieve this goal. One of the most promising approaches is the use of double-stranded DNA (dsDNA) to specifically target and inhibit the expression of genes.
DsDNA is a molecule that consists of two complementary strands of DNA that are bound together by hydrogen bonds. This structure allows dsDNA to bind to specific regions of the genome, which can be used to target and inhibit the expression of specific genes. This approach has several advantages over other methods of gene inhibition, including specificity, efficiency, and ease of use.
The market for methods, compositions, and compounds for the specific inhibition by dsDNA of gene expression is driven by the increasing demand for new and effective treatments for diseases that are caused by abnormal gene expression. The global market for gene therapy is expected to reach $13.3 billion by 2024, with a compound annual growth rate of 33.3% from 2019 to 2024.
Several companies are currently developing methods and compounds for the specific inhibition by dsDNA of gene expression. These include CRISPR Therapeutics, Sangamo Therapeutics, and Editas Medicine, among others. These companies are using a variety of approaches, including the use of RNA interference (RNAi), antisense oligonucleotides (ASOs), and gene editing technologies such as CRISPR/Cas9.
The market for methods, compositions, and compounds for the specific inhibition by dsDNA of gene expression is expected to continue to grow in the coming years, driven by the increasing demand for new and effective treatments for diseases that are caused by abnormal gene expression. This market represents a significant opportunity for biotechnology companies that are developing innovative approaches to gene therapy.
In conclusion, the market for methods, compositions, and compounds for the specific inhibition by dsDNA of gene expression is a rapidly growing field in the biotechnology industry. This market is driven by the increasing demand for new and effective treatments for diseases that are caused by abnormal gene expression. As the field continues to evolve, it is likely that new and innovative approaches will be developed, leading to the development of new treatments for a wide range of diseases.
The Integrated DNA Technologies Inc, City of Hope invention works as follows
The invention is directed at compositions and techniques for selectively reducing expression of a target gene’s gene product in a cell as well as treating diseases that are caused by gene expression. The invention is more specifically directed at compositions that contain double-stranded DNA (?dsRNA?) The invention is directed to compositions that contain double stranded RNA (?dsRNA? ), as well as methods of preparing them. These compositions and methods are capable of reducing expression of target gene in eukaryotic cell. The dsRNA consists of a first oligonucleotide that ranges between 25 to 30 nucleotides and a second sequence that, under biological conditions, anneals with the first sequence. A region of a dsRNA sequence with a length of 19 nucleotides or more is complementary enough to a target RNA nucleotide to trigger destruction by the RNAi mechanism.
Background for Methods, compositions and compounds for the specific inhibition by double-stranded DNA of gene expression
Suppression of gene expression by double-stranded RNA (dsRNA) has been demonstrated in a variety of systems including plants (post-transcriptional gene suppression) (Napoli et al., 1990), fungi (quelling) (Romano and Marcino, 1992), and nematodes (RNA interference) (Fire et al., 1998). Double-stranded (dsRNA), which is more stable than ssRNA, has been shown to suppress gene expression in a variety of systems including plants (post-transcriptional gene suppression) (Napoli et al., 1990), fungi (quelling) (Romano and Marcino 1992), and nematodes (RNA interference). This difference is more pronounced within the intracellular environment. (Raemdonck and colleagues, 2006). Unmodified siRNAs degrade rapidly in serum which is a nuclease-rich environment. Chemical modification of siRNA can improve potency in vitro as well as in vivo. Over the last 20 years, extensive medicinal chemistry was done for applications that use synthetic nucleic acid for experimental or therapeutic purposes in vivo. This includes the antisense field and the ribozyme field. Hundreds of compounds were tested to find modifications that improved nuclease stabilization, increased binding affinity and sometimes even improved pharmacodynamics of synthetic nucleic acid (Matteucci 1997; Manoharan 2002; Kurreck 2003). Many of these modifications were tested and have been found to be useful as modifiers in 21mer siRNAs. Many reviews summarize recent experience with 21mer RNAs and chemical modification (Zhang, Sipa and Nawrot, 2006, Rana, 2007, Rana et al.). “Modification patterns haven’t been optimized or tested for use with longer RNAs such as Dicer substrate siRNAs.
Early attempts to suppress gene transcription using long dsRNAs failed in mammalian system due to activation interferon pathways which do not exist in lower animals. DsRNAs trigger interferon responses (Stark and al., 1998). Specifically, the protein kinase PKR, which is activated by dsRNAs longer than 30 bp (Manche et. al.,1992), results in phosphorylation eIF2’s translation initiation factor. This leads to the arrest of protein synthesis, and activation 2?5-oligoadenolate synthetase (OAS) which results in RNA degradation.
In Drosophila cell extracts and cells, longer dsRNAs were effective in RNA interfering, while shorter dsRNAs did not (Tuschl, 1999). The active molecule is long double-stranded DNA, but it’s not the effector. Long dsRNAs can be degraded into short duplexes of 21-23 bp with 2-base 3-overhangs by Dicer, an RNase III enzyme (Bernstein, et. al. 2001). The short RNA duplexes are called siRNAs and they direct the RNAi reaction in vivo. Transfection of chemically synthesized short siRNA duplexes allows RNAi methods in mammalian cell to suppress gene expression without triggering unwanted Interferon responses. The antisense strand in the siRNA duplex acts as a sequence specific guide to direct the activity of an endoribonuclease enzyme function within the RNA induced silence complex (RISC). This leads to the degradation of target mRNA.
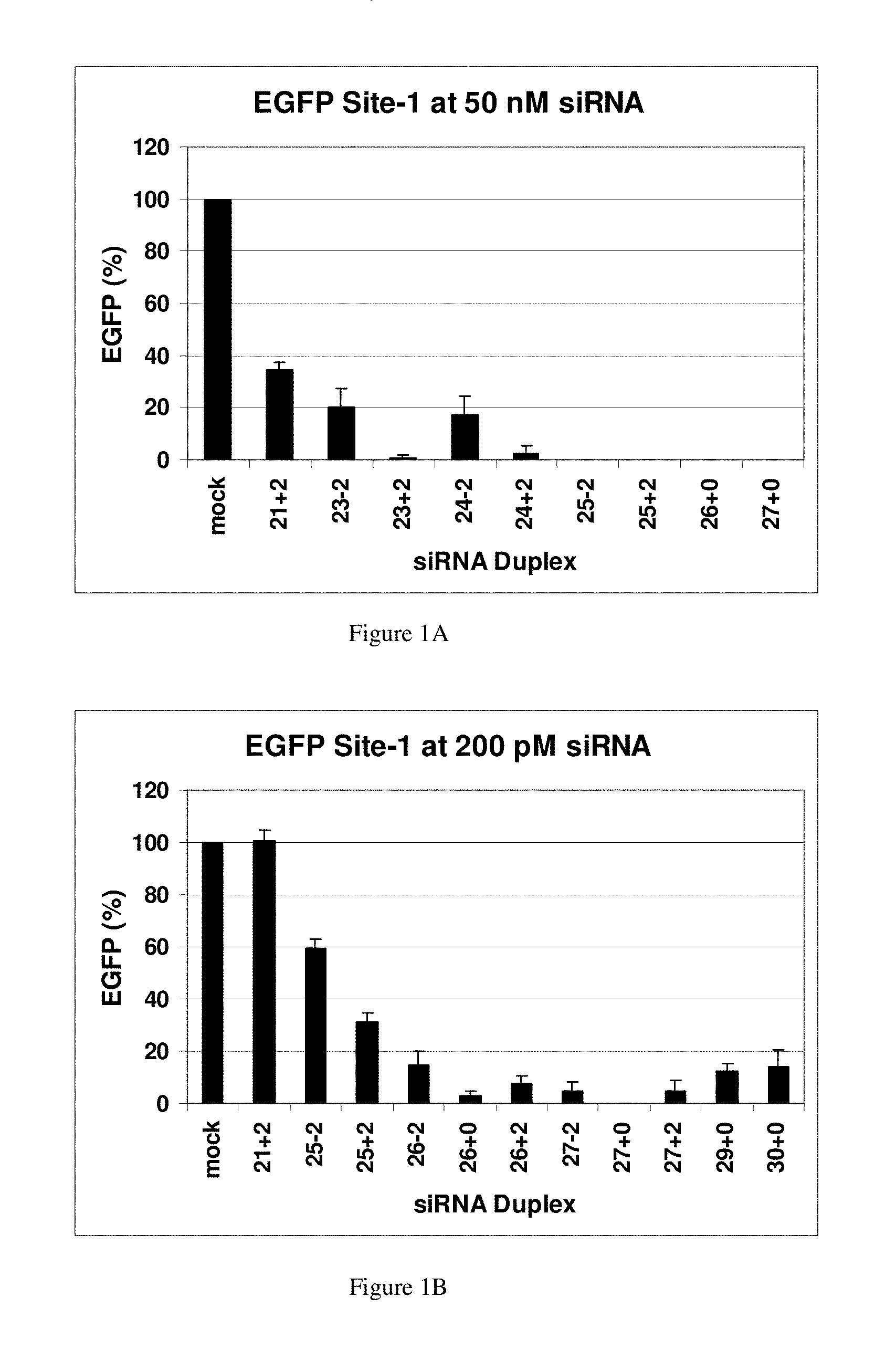
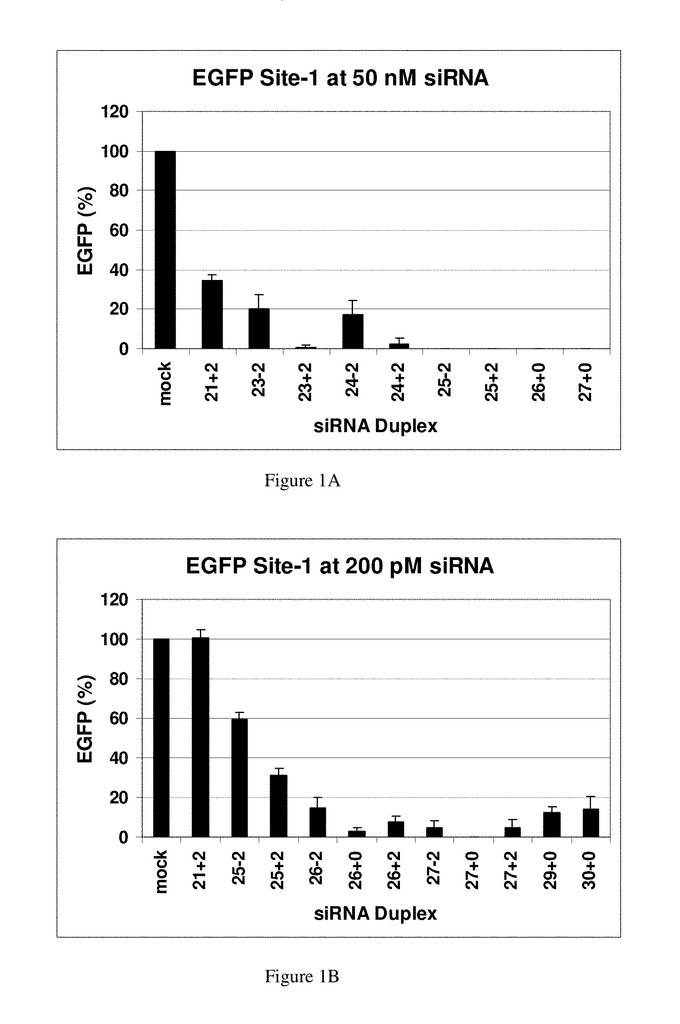
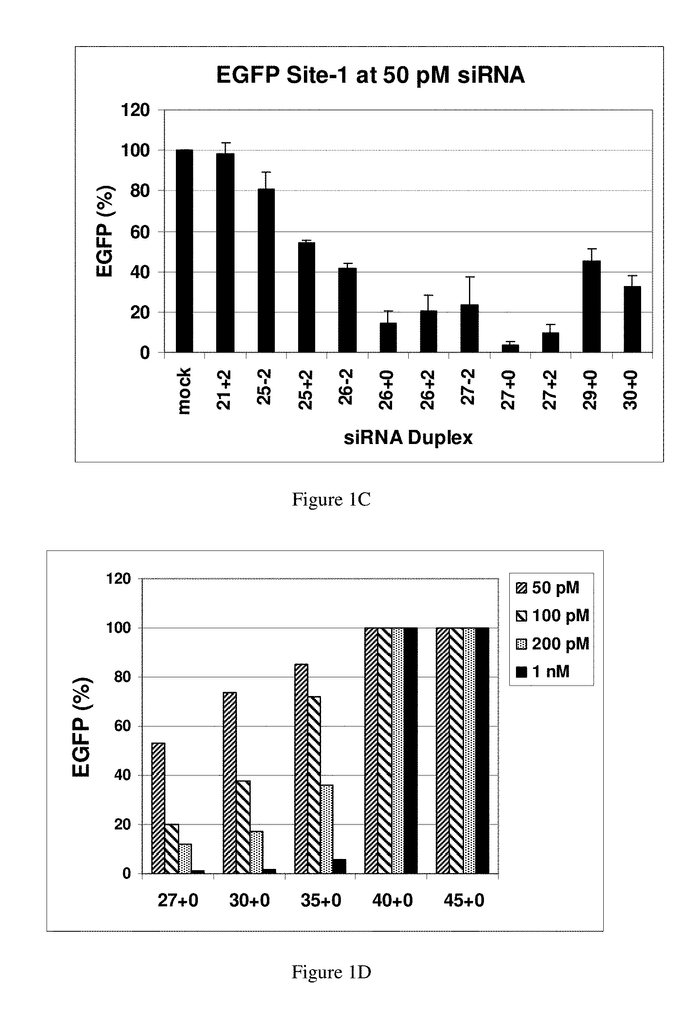
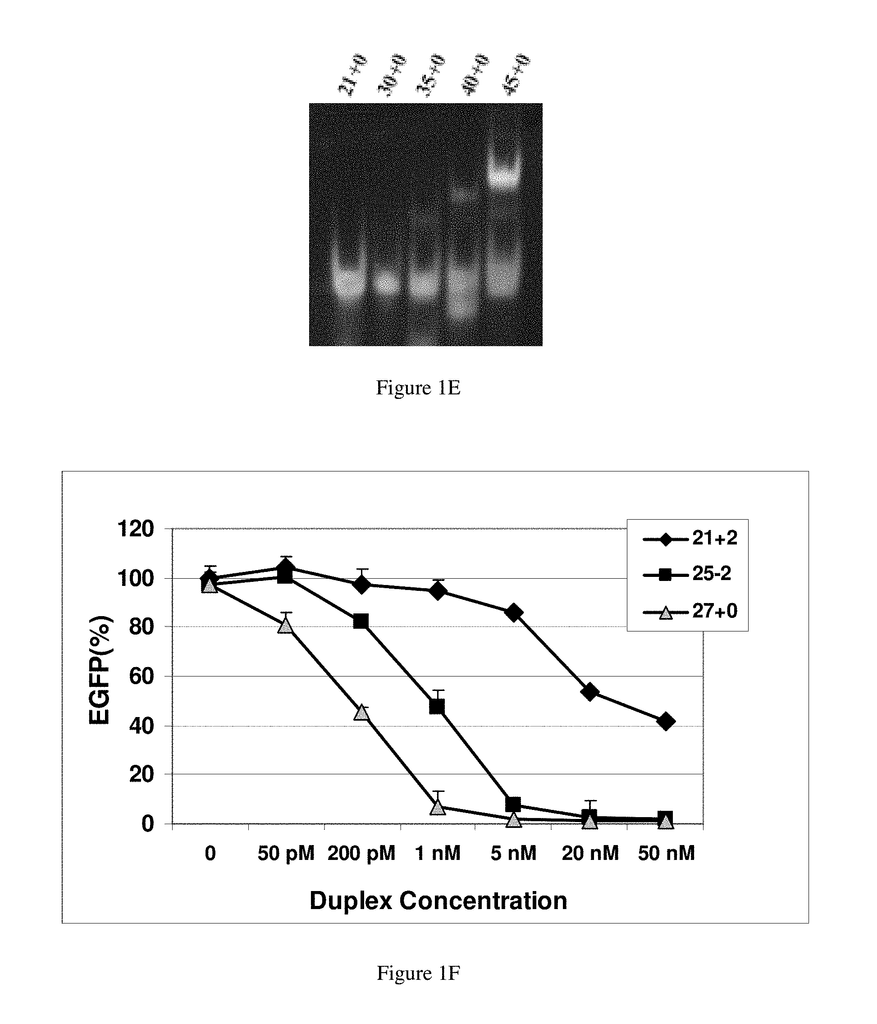
In studying the size limit for RNAi, in Drosophila extracts, in vitro a lower threshold was established of around 38 bp for activation of RNAi using exogenously-supplied double-stranded RNA, and duplexes 36, 30 and 29 bp in length (Elbashir et.al., 2001b). The 30-base RNAs did not cleave into 21-23 base siRNAs, and were therefore deemed inactive to be used in RNAi. (Elbashir et. al. 2001b). The same group continued to map the structural requirements for chemically synthesized RNAs to function in RNAi pathways. RNA duplexes with 2-base 3-overhangs and 21-bp were the most effective. Duplexes 20-22-bp and 23-bp were slightly less potent but still resulted in RNAi-mediated mRNA degrada-tion, while 24-25-bp were inactive.
Some of the conclusions from these earlier studies could be specific to Drosophila. Some investigators have shown that siRNAs longer than 21-23 bp can be effective in human cells. Duplexes between 21-23 bp have been found to be more effective and are now the preferred design (Caplen, et. al., 2001). Chemically synthesized duplex RNAs mimicking the natural products resulting from Dicer degradation were found to be preferred for use in RNAi. Researchers studying the size limits of RNAi for Caenorhabditis Elegans approached this problem in the opposite direction. They found that a microinjected RNA duplex 26-bp could suppress gene expression but it required a 250-fold increase in concentration when compared to an 81-bp.
Despite recent attention on RNAi, the field remains in its early stages of development. Some siRNA molecules can’t target the destruction of complementary RNAs within a cell. As a consequence, complex rules were developed to design RNAi molecules. The experts in this field expect to test many siRNA molecules before finding the right composition. (Ji, et al. 2003) Some experts pool together several siRNA preparations to increase their chances of silencing a target in a single experiment. These pools contain up to 20 nM, or even more, of siRNA duplexes (Ji, 2003). This is despite the fact a siRNA can be effective at concentrations as low as 1 nM (Holen, 2002). This technique may produce artifacts due to interactions between siRNA sequences and other cellular RNAs. Off target effects may occur when RNAi oligonucleotides are homologous to unintended targets, or when the RISC-complex incorporates an unintended strand of RNAi duplex. These effects are generally more pronounced with higher concentrations of RNAi-duplexes. (Scherer et al., 2003)
In addition, RNAi treatments are only effective for a maximum of 4 days” (Holen et. al.,2002). Researchers must perform siRNA experiments within 2 to 3 days after transfection of an siRNA duplex, or use plasmids or viral expression vectors for longer-term silencing.
Nucleases are responsible for the degradation of siRNAs, which is a major factor in the inhibition of the siRNA effect. The primary nuclease in serum is a 3?-exonuclease. Modification of the 3? ends of antisense DNA is essential to prevent degradation. ERI-1, a nuclease of the RNase-T subfamily with 3??5? has been identified. Exonuclease is a key enzyme in the regulation and degradation siRNAs. (Kennedy, 2004; Hong, 2005). This gene, also known as Thex1 in mice (NM_02067) or THEX1 in humans (NM_153332), is involved in the degradation of histones mRNA. It also mediates the degradation of 3? overhangs in siRNAs but does not degrade RNA duplex (Yang, et. al., 2006. It is reasonable to expect 3?-end stabilization of siRNAs to improve stability.
XRN1(NM_019001), is a 5??3?? exonuclease that resides in P-bodies and has been implicated in degradation of mRNA targeted by miRNA (Rehwinkel et al., 2005). It may also be responsible for completing the degradation initiated by internal cleavage as directed by rRNA. “XRN1 (NM_019001) is a 5??3? XRN2 is a 5??3?? exonuclease. Exonuclease involved in the processing of nuclear RNA. “Although not implicated currently in the degradation or processing siRNAs and microRNAs, both of these nucleases can degrade RNAs. They may also be important to take into consideration.
RNase A is the major endonuclease in mammals. It degrades RNAs. It is specific to ssRNA, and it cleaves the 3?-end pyrimidine base. After incubation with serum, mass spectrometry can detect SiRNA degradation products consistent RNase A. The 3?-overhangs increase the susceptibility to RNase degradation of siRNAs. The degradation of siRNAs is reduced when RNase A in serum is depleted. This degradation shows some sequence preference, and it’s worse for sequences with poly A/U on the ends. It is possible that the lower stability regions may “breathe” This suggests that lower stability regions of the duplex may?breathe?
In 21mers phosphorothioate and boranophosphate modification directly stabilize the internucleoside-phosphate linkage. Boranophosphate-modified RNAs have a high nuclease resistance, are potent silencing agents and are relatively safe. Boranophosphate-modified RNAs are not made using chemical synthesis, but instead by in vitro transcription. (Hall et. al. 2004 and Hall, et. al. 2006). Standard chemical synthesis can easily place PS modifications in any desired position on the RNA duplex. Most investigators recommend limiting the PS modification in siRNAs and favoring 3?-ends for protection against nucleases (Harborth, 2003, Chiu, Rana, Braasch, et. al. 2003, Amarzguioui, et. al. 2003). The PS modification is compatible with potent activity of RNAi, but sugar modifications, such as 2?-O methyl RNA, may be superior.
A number of substitutions may be made at the 2? position of the ribose, which can improve nuclease resistant and increase duplex stability. The 2?-O methyl modification is found in transfer RNAs, ribosomal and mammalian transcripts. The 2?O-methyl modification is well-known in siRNAs, but it is the exact position of the modified bases in the duplex that is crucial to maintain potency. Complete substitution of 2′-O-methyl-RNA with RNA will result in inactivation of the siRNA. “For example, a pattern involving alternating 2?O-methyl bases has potency equal to unmodified DNA and is stable in serum. (Choung, Czauderna, 2003; Choung, Czauderna, 2006).
The 2?F modification can also be used in combination with the 2?O-methyl modification. 2?F purines can also be obtained and used. These heavily modified duplexes can trigger RNAi effectively in vitro (Allerson et. al. 2005; Prakash et. al. 2005; Kraynack & Baker, 2006), and they can also improve performance and prolong the duration of action in vivo. Allerson teaches a nuclease-stable, blunt duplex that contains alternative 2??-F or 2??-O?-Me bases. This design uses the same pattern as Czauderna but converts the remaining RNA to 2?F modified bases. Morrissey used a nuclease-resistant siRNA that was highly potent. This duplex contains DNA, RNA inverted abasics and a 3′-terminal PS internucleoside. Although extensive modification can have certain advantages, a more limited modification to the duplex may also improve its in vivo performance. It is simpler and cheaper to manufacture. Soutschek et al. In vivo, the duplex was mainly RNA with 2?-O?-Me RNA base and limited 3??-terminal internucleoside links.
Locked Nucleic Acids (LNAs), a class of 2?-modification, can be used to stabilise siRNAs. LNA incorporation patterns that maintain potency are more limited than 2?O-methyl bases or 2?F bases. Limited modification is therefore preferred (Braasch, Grunweller, Elmen, 2005). “Even with limited incorporation of LNA modifications, siRNA performance can be improved in vivo, and off-target effect profiles may also be altered or improved (Mook, et. al., 2007.
Synthetic Nucleic Acids introduced into live cells or animals can be identified as ‘foreign’ Immune response Immune stimulation is a class of effects that can have a dramatic impact on experimental results, and may even cause cell death. The innate immunity system contains a number of receptor molecules which interact specifically with DNA andRNA to mediate the responses. Some of these receptors are located in cytoplasm, while others reside in endosomes. The delivery of siRNAs using cationic liposomes or lipids exposes siRNAs to both the cytoplasmic compartment and the endosomal compartment, increasing the risk of triggering an IFN type 1 response in vitro and also in vivo. (Morrissey and al. 2005b; Sioud & Sorensen 2003; Sioud 2005; Ma and al. 2005). RNAs transcribed in the cell are not immunogenic (Robbins and Sorensen, 2006). Synthetic RNAs, which are immunogenic, can be introduced into cells mechanically, even when they are delivered by lipid-based techniques (Heidel and al. 2004). Lipid-based delivery methods, however, are effective, convenient, and widely used. A general strategy is required to prevent immune reactions, particularly for in vivo applications where all cell types present are present and there is a high risk of generating a response. Chemically modified RNAs could solve many or all of these issues.
The receptors of innate immunity are able to distinguish between the presence and absence of certain base modification motifs that are more common in mammalian than prokaryotic DNA. Examples of these modified bases include pseudouridine, 2?-O?-methyl, N6-methyl, and N6-methyl A. Inclusion of these residues can help to evade immune recognition (Kariko, 2005). When administered intravenously to mice, extensive 2?-modification can be used to block the immune response. (Morrissey and al. 2005b). It is not necessary to modify the sequence extensively to avoid immune detection. Substituting two 2?-Omethyl bases into a single duplex of siRNA can be enough to block type 1 IFN responses both in vitro as well as in vivo. Modified U and G bases work best (Judge and al., 2006). Selective incorporation of 2-O-methyl base can also reduce the magnitude off-target effects. (Jackson and al., 2006.) The use of 2?-O methyl bases for siRNAs that are intended for in-vivo application is a good way to block immune responses. It also has the benefit of increasing nuclease stabilty and reducing off-target effects.
Although immune stimulation can result in cell death, assessing the viability of cells is not a reliable method for monitoring IFN response induction. IFN responses are possible without cell death. Cell death can also result from knockdown of the target gene in the absence IFN activation (for instance, if that gene is vital for cell viability). The cytokines that are relevant can be measured directly in the culture medium. A variety of commercial kits make it easy to perform such tests. Testing levels of IFN?, TNF? and IL-6 4 and 24 hours after transfection are usually enough to screen for a number of immune effector molecules. Include a “transfection reagent-only control” As cationic lipoproteins can trigger immune reactions in cells without any nucleic acids, it is important to include a?transfection reagent only control? Cell culture experiments should include controls for IFN pathway activation. When administering nucleic acid in vivo (where the risk of IFN response is greatest), it is important to test for immune stimuli.
The modifications should not reduce potency, 2) shouldn’t interfere with Dicer processing; 3) improve stability in biological fluids (reduce nuclease sensitivity); 4) block or evade detection by the innate immune system; 5) shouldn’t be toxic and 6) shouldn?t increase cost or impact ease of manufacturing. The modifications should 1) not reduce potency, 2) not interfere with Dicer process; 3) improve stability in bio fluids (reduces nuclease sensitivities); 4) not be toxic and 5) not increase the cost or impact on ease of manufacture.
The invention provides compositions that are useful for RNAi to inhibit gene expression, and methods for using them. The invention also provides RNAi formulations and methods that are designed to maximize potency and enhance Dicer processing. They are also stable, evade the immune system, and not toxic. The invention can be adapted to high-throughput small-scale synthesis for research purposes as well as for large-scale manufacturing for therapeutic applications. The description of this invention will reveal these and other benefits of the invention as well as inventive features.
The present invention relates to compositions containing double-stranded (dsRNA) RNA and methods for preparing them, which are capable of reducing the expression of target genes in eukaryotic cells. The invention relates to compositions that contain double stranded RNA (?dsRNA? ), as well as methods of preparing them. These compositions and methods are capable of reducing expression of target gene in eukaryotic cell. The invention is directed more specifically to Dicer substrates RNAs that have been functionally enhanced.
In a first instance, the invention provides novel compositions that are suitable for RNA interfering (RNAi).” The compositions contain dsRNA, which is a pre-molecule. i.e. the dsRNA from the present invention is then processed in vivo into an active siRNA. Dicer converts the dsRNA into an active siRNA that is incorporated in the RISC complex to RNA interfere with a target gene. The precursor molecule can also be called a RNAi molecule precursor herein.
Click here to view the patent on Google Patents.